The ability to resume an analysis (i.e. caching) is one of the core strengths of Nextflow. When developing pipelines, this allows us to avoid re-running unchanged processes by simply appending -resume to the nextflow run command. Sometimes, tasks may be repeated for reasons that are unclear. In these cases it can help to look into the caching mechanism, to understand why a specific process was re-run.
We have previously written about Nextflow’s resume functionality as well as some troubleshooting strategies to gain more insights on the caching behavior.
In this post, we will take a more hands-on approach and highlight some strategies which we can use to understand what is causing a particular process (or processes) to re-run, instead of using the cache from previous runs of the pipeline. To demonstrate the process, we will introduce a minor change into one of the process definitions in the the nextflow-io/rnaseq-nf pipeline and investigate how it affects the overall caching behavior when compared to the initial execution of the pipeline.
First, we clone the nextflow-io/rnaseq-nf pipeline locally:
$ git clone https://github.com/nextflow-io/rnaseq-nf
$ cd rnaseq-nf
In the examples below, we have used Nextflow v22.10.0, Docker v20.10.8 and Java v17 LTS on MacOS.
The flowchart below can help in understanding the design of the pipeline and the dependencies between the various tasks.
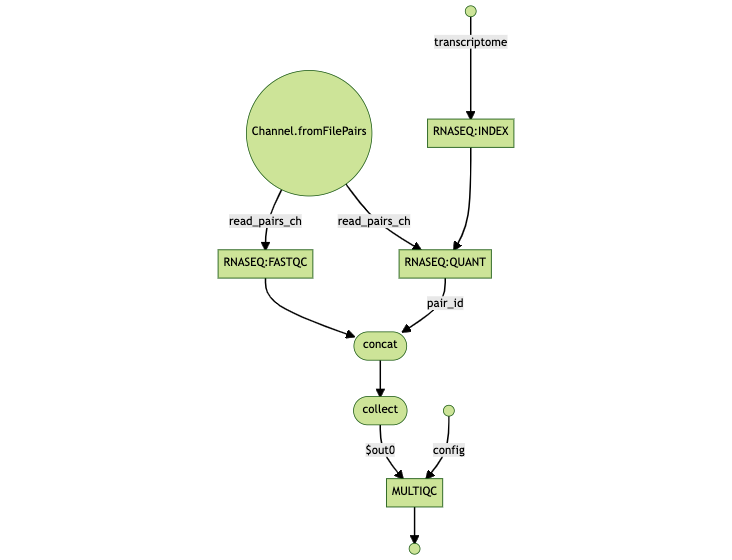
As a reminder, Nextflow generates a unique task hash, e.g. 22/7548fa… for each task in a workflow. The hash takes into account the complete file path, the last modified timestamp, container ID, content of script directive among other factors. If any of these change, the task will be re-executed. Nextflow maintains a list of task hashes for caching and traceability purposes. You can learn more about task hashes in the article Troubleshooting Nextflow resume.
To have something to compare to, we first need to generate the initial hashes for the unchanged processes in the pipeline. We save these in a file called fresh_run.log and use them later on as “ground-truth” for the analysis. In order to save the process hashes we use the -dump-hashes flag, which prints them to the log.
TIP: We rely upon the -log option in the nextflow command line interface to be able to supply a custom log file name instead of the default .nextflow.log.
$ nextflow -log fresh_run.log run ./main.nf -profile docker -dump-hashes
[...truncated…]
executor > local (4)
[d5/57c2bb] process > RNASEQ:INDEX (ggal_1_48850000_49020000) [100%] 1 of 1 ✔
[25/433b23] process > RNASEQ:FASTQC (FASTQC on ggal_gut) [100%] 1 of 1 ✔
[03/23372f] process > RNASEQ:QUANT (ggal_gut) [100%] 1 of 1 ✔
[38/712d21] process > MULTIQC [100%] 1 of 1 ✔
[...truncated…]
FastQC processAfter the initial run of the pipeline, we introduce a change in the fastqc.nf module, hard coding the number of threads which should be used to run the FASTQC process via Nextflow’s cpus directive.
Here’s the output of git diff on the contents of modules/fastqc/main.nf file:
--- a/modules/fastqc/main.nf
+++ b/modules/fastqc/main.nf
@@ -4,6 +4,7 @@ process FASTQC {
tag "FASTQC on $sample_id"
conda 'bioconda::fastqc=0.11.9'
publishDir params.outdir, mode:'copy'
+ cpus 2
input:
tuple val(sample_id), path(reads)
@@ -13,6 +14,6 @@ process FASTQC {
script:
"""
- fastqc.sh "$sample_id" "$reads"
+ fastqc.sh "$sample_id" "$reads" -t ${task.cpus}
"""
}
Next, we run the pipeline again with the -resume option, which instructs Nextflow to rely upon the cached results from the previous run and only run the parts of the pipeline which have changed. As before, we instruct Nextflow to dump the process hashes, this time in a file called resumed_run.log.
$ nextflow -log resumed_run.log run ./main.nf -profile docker -dump-hashes -resume
[...truncated…]
executor > local
[d5/57c2bb] process > RNASEQ:INDEX (ggal_1_48850000_49020000) [100%] 1 of 1, cached: 1 ✔
[55/15b609] process > RNASEQ:FASTQC (FASTQC on ggal_gut) [100%] 1 of 1 ✔
[03/23372f] process > RNASEQ:QUANT (ggal_gut) [100%] 1 of 1, cached: 1 ✔
[f3/f1ccb4] process > MULTIQC [100%] 1 of 1 ✔
[...truncated…]
From the summary of the command line output above, we can see that the RNASEQ:FASTQC (FASTQC on ggal_gut) and MULTIQC processes were re-run while the others were cached. To understand why, we can examine the hashes generated by the processes from the logs of the fresh_run and resumed_run.
For the analysis, we need to keep in mind that:
The time-stamps are expected to differ and can be safely ignored to narrow down the grep pattern to the Nextflow TaskProcessor class.
The order of the log entries isn’t fixed, due to the nature of the underlying parallel computation dataflow model used by Nextflow. For example, in our example below, FASTQC ran first in fresh_run.log but wasn’t the first logged process in resumed_run.log.
We can use standard Unix tools like grep, cut and sort to address these points and filter out the relevant information:
grep to isolate log entries with cache hash stringcut -d ‘-’ -f 3cut -d ';' -f 1sort to have a standard order before comparisontee to print the resultant strings to the terminal and simultaneously save to a fileNow, let’s apply these transformations to the fresh_run.log as well as resumed_run.log entries.
fresh_run.log$ cat ./fresh_run.log | grep 'INFO.*TaskProcessor.*cache hash' | cut -d '-' -f 3 | cut -d ';' -f 1 | sort | tee ./fresh_run.tasks.log
[MULTIQC] cache hash: 167d7b39f7efdfc49b6ff773f081daef
[RNASEQ:FASTQC (FASTQC on ggal_gut)] cache hash: 47e8c58d92dbaafba3c2ccc4f89f53a4
[RNASEQ:INDEX (ggal_1_48850000_49020000)] cache hash: ac8be293e1d57f3616cdd0adce34af6f
[RNASEQ:QUANT (ggal_gut)] cache hash: d8b88e3979ff9fe4bf64b4e1bfaf4038
resumed_run.log$ cat ./resumed_run.log | grep 'INFO.*TaskProcessor.*cache hash' | cut -d '-' -f 3 | cut -d ';' -f 1 | sort | tee ./resumed_run.tasks.log
[MULTIQC] cache hash: d3f200c56cf00b223282f12f06ae8586
[RNASEQ:FASTQC (FASTQC on ggal_gut)] cache hash: 92478eeb3b0ff210ebe5a4f3d99aed2d
[RNASEQ:INDEX (ggal_1_48850000_49020000)] cache hash: ac8be293e1d57f3616cdd0adce34af6f
[RNASEQ:QUANT (ggal_gut)] cache hash: d8b88e3979ff9fe4bf64b4e1bfaf4038
Computing a hash is a multi-step process and various factors contribute to it such as the inputs of the process, platform, time-stamps of the input files and more ( as explained in Demystifying Nextflow resume blog post) . The change we made in the task level CPUs directive and script section of the FASTQC process triggered a re-computation of hashes:
--- ./fresh_run.tasks.log
+++ ./resumed_run.tasks.log
@@ -1,4 +1,4 @@
- [MULTIQC] cache hash: dccabcd012ad86e1a2668e866c120534
- [RNASEQ:FASTQC (FASTQC on ggal_gut)] cache hash: 94be8c84f4bed57252985e6813bec401
+ [MULTIQC] cache hash: c5a63560338596282682cc04ff97e436
+ [RNASEQ:FASTQC (FASTQC on ggal_gut)] cache hash: 54aa712db7c8248e7f31d5fb6535ff9d
[RNASEQ:INDEX (ggal_1_48850000_49020000)] cache hash: 356aaa7524fb071f258480ba07c67b3c
[RNASEQ:QUANT (ggal_gut)] cache hash: 169ced0fc4b047eaf91cd31620b22540
Even though we only introduced changes in FASTQC, the MULTIQC process was re-run since it relies upon the output of the FASTQC process. Any task that has its cache hash invalidated triggers a rerun of all downstream steps:
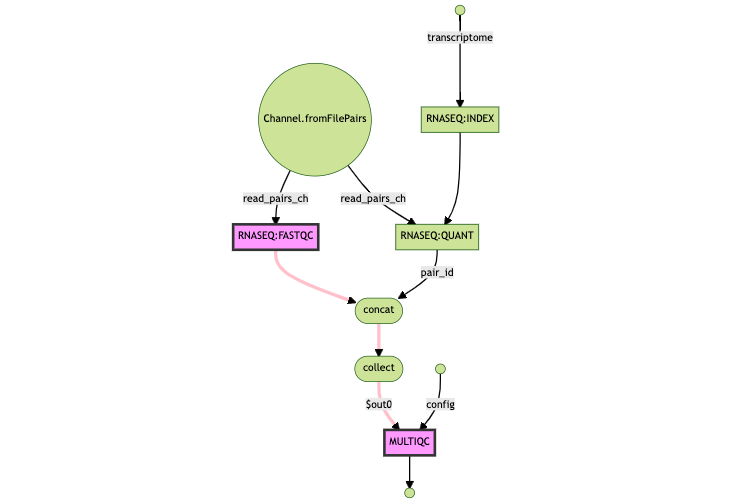
FASTQC was re-runWe can see the full list of FASTQC process hashes within the fresh_run.log file
[...truncated…]
Nov-03 20:19:13.827 [Actor Thread 6] INFO nextflow.processor.TaskProcessor - [RNASEQ:FASTQC (FASTQC on ggal_gut)] cache hash: 54aa712db7c8248e7f31d5fb6535ff9d; mode: STANDARD; entries:
1a0e496fef579b22998f099981b494f9 [java.util.UUID] a11bf24f-638a-42d6-8b50-48d3be637d54
195c7faea83c75f2340eb710d8486d2a [java.lang.String] RNASEQ:FASTQC
2bea0eee5e384bd6082a173772e939eb [java.lang.String] """
fastqc.sh "$sample_id" "$reads" -t ${task.cpus}
"""
8e58c0cec3bde124d5d932c7f1579395 [java.lang.String] quay.io/nextflow/rnaseq-nf:v1.1
7ec7cbd71ff757f5fcdbaa760c9ce6de [java.lang.String] sample_id
16b4905b1545252eb7cbfe7b2a20d03d [java.lang.String] ggal_gut
553096c532e666fb42214fdf0520fe4a [java.lang.String] reads
6a5d50e32fdb3261e3700a30ad257ff9 [nextflow.util.ArrayBag] [FileHolder(sourceObj:/home/abhinav/rnaseq-nf/data/ggal/ggal_gut_1.fq, storePath:/home/abhinav/rnaseq-nf/data/ggal/ggal_gut_1.fq, stageName:ggal_gut_1.fq), FileHolder(sourceObj:/home/abhinav/rnaseq-nf/data/ggal/ggal_gut_2.fq, storePath:/home/abhinav/rnaseq-nf/data/ggal/ggal_gut_2.fq, stageName:ggal_gut_2.fq)]
4f9d4b0d22865056c37fb6d9c2a04a67 [java.lang.String] $
16fe7483905cce7a85670e43e4678877 [java.lang.Boolean] true
80a8708c1f85f9e53796b84bd83471d3 [java.util.HashMap$EntrySet] [task.cpus=2]
f46c56757169dad5c65708a8f892f414 [sun.nio.fs.UnixPath] /home/abhinav/rnaseq-nf/bin/fastqc.sh
[...truncated…]
When we isolate and compare the log entries for FASTQC between fresh_run.log and resumed_run.log, we see the following diff:
--- ./fresh_run.fastqc.log
+++ ./resumed_run.fastqc.log
@@ -1,8 +1,8 @@
-INFO nextflow.processor.TaskProcessor - [RNASEQ:FASTQC (FASTQC on ggal_gut)] cache hash: 94be8c84f4bed57252985e6813bec401; mode: STANDARD; entries:
+INFO nextflow.processor.TaskProcessor - [RNASEQ:FASTQC (FASTQC on ggal_gut)] cache hash: 54aa712db7c8248e7f31d5fb6535ff9d; mode: STANDARD; entries:
1a0e496fef579b22998f099981b494f9 [java.util.UUID] a11bf24f-638a-42d6-8b50-48d3be637d54
195c7faea83c75f2340eb710d8486d2a [java.lang.String] RNASEQ:FASTQC
- 43e5a23fc27129f92a6c010823d8909b [java.lang.String] """
- fastqc.sh "$sample_id" "$reads"
+ 2bea0eee5e384bd6082a173772e939eb [java.lang.String] """
+ fastqc.sh "$sample_id" "$reads" -t ${task.cpus}
Observations from the diff:
$task.cpus part of the command.resumed_run.log showing that the content of the process level directive cpus has been added.In other words, the diff from log files is confirming our edits.
MULTIQC was re-runNow, we apply the same analysis technique for the MULTIQC process in both log files:
--- ./fresh_run.multiqc.log
+++ ./resumed_run.multiqc.log
@@ -1,4 +1,4 @@
-INFO nextflow.processor.TaskProcessor - [MULTIQC] cache hash: dccabcd012ad86e1a2668e866c120534; mode: STANDARD; entries:
+INFO nextflow.processor.TaskProcessor - [MULTIQC] cache hash: c5a63560338596282682cc04ff97e436; mode: STANDARD; entries:
1a0e496fef579b22998f099981b494f9 [java.util.UUID] a11bf24f-638a-42d6-8b50-48d3be637d54
cd584abbdbee0d2cfc4361ee2a3fd44b [java.lang.String] MULTIQC
56bfc44d4ed5c943f30ec98b22904eec [java.lang.String] """
@@ -9,8 +9,9 @@
8e58c0cec3bde124d5d932c7f1579395 [java.lang.String] quay.io/nextflow/rnaseq-nf:v1.1
14ca61f10a641915b8c71066de5892e1 [java.lang.String] *
- cd0e6f1a382f11f25d5cef85bd87c3f4 [nextflow.util.ArrayBag] [FileHolder(sourceObj:/home/abhinav/rnaseq-nf/work/03/23372f156e80deb4d7183c5f509274/ggal_gut, storePath:/home/abhinav/rnaseq-nf/work/03/23372f156e80deb4d7183c5f509274/ggal_gut, stageName:ggal_gut), FileHolder(sourceObj:/home/abhinav/rnaseq-nf/work/25/433b23af9e98294becade95db6bd76/fastqc_ggal_gut_logs, storePath:/home/abhinav/rnaseq-nf/work/25/433b23af9e98294becade95db6bd76/fastqc_ggal_gut_logs, stageName:fastqc_ggal_gut_logs)]
+ 18966b473f7bdb07f4f7f4c8445be1f5 [nextflow.util.ArrayBag] [FileHolder(sourceObj:/home/abhinav/rnaseq-nf/work/03/23372f156e80deb4d7183c5f509274/ggal_gut, storePath:/home/abhinav/rnaseq-nf/work/03/23372f156e80deb4d7183c5f509274/ggal_gut, stageName:ggal_gut), FileHolder(sourceObj:/home/abhinav/rnaseq-nf/work/55/15b60995682daf79ecb64bcbb8e44e/fastqc_ggal_gut_logs, storePath:/home/abhinav/rnaseq-nf/work/55/15b60995682daf79ecb64bcbb8e44e/fastqc_ggal_gut_logs, stageName:fastqc_ggal_gut_logs)]
d271b8ef022bbb0126423bf5796c9440 [java.lang.String] config
5a07367a32cd1696f0f0054ee1f60e8b [nextflow.util.ArrayBag] [FileHolder(sourceObj:/home/abhinav/rnaseq-nf/multiqc, storePath:/home/abhinav/rnaseq-nf/multiqc, stageName:multiqc)]
4f9d4b0d22865056c37fb6d9c2a04a67 [java.lang.String] $
16fe7483905cce7a85670e43e4678877 [java.lang.Boolean] true
Here, the highlighted diffs show the directory of the input files, changing as a result of FASTQC being re-run; as a result MULTIQC has a new hash and has to be re-run as well.
Debugging the caching behavior of a pipeline can be tricky, however a systematic analysis can help to uncover what is causing a particular process to be re-run.
When analyzing large datasets, it may be worth using the -dump-hashes option by default for all pipeline runs, avoiding needing to run the pipeline again to obtain the hashes in the log file in case of problems.
While this process works, it is not trivial. We would love to see some community-driven tooling for a better cache-debugging experience for Nextflow, perhaps an nf-cache plugin? Stay tuned for an upcoming blog post describing how to extend and add new functionality to Nextflow using plugins.